Influence Diagram¶
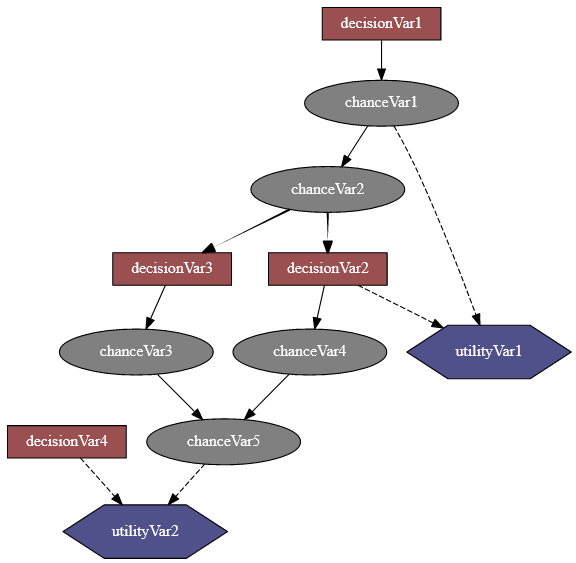
An influence diagram is a compact graphical and mathematical representation of a decision situation. It is a generalization of a Bayesian network, in which not only probabilistic inference problems but also decision making problems (following the maximum expected utility criterion) can be modeled and solved. It includes 3 types of nodes : action, decision and utility nodes (from wikipedia).
PyAgrum’s so-called influence diagram represents both influence diagrams and LIMIDs. The way to enforce that such a model represent an influence diagram and not a LIMID belongs to the inference engine.
Tutorial
Reference
Model¶
-
class
pyAgrum.InfluenceDiagram(*args)¶ InfluenceDiagram represents an Influence Diagram.
- InfluenceDiagram() -> InfluenceDiagram
- default constructor
- InfluenceDiagram(source) -> InfluenceDiagram
- Parameters:
- source (pyAgrum.InfluenceDiagram) – the InfluenceDiagram to copy
-
add(self, variable, id=0)¶ Add a chance variable, it’s associate node and it’s CPT.
The id of the new variable is automatically generated.
Parameters: - variable (pyAgrum.DiscreteVariable) – The variable added by copy.
- id (int) – The chosen id. If 0, the NodeGraphPart will choose.
Warning
give an id (not 0) should be reserved for rare and specific situations !!!
Returns: the id of the added variable. Return type: int Raises: gum.DuplicateElement– If id(<>0) is already used
-
addArc(self, tail, head)¶ addArc(self, tail, head)
Add an arc in the ID, and update diagram’s potential nodes cpt if necessary.
Parameters: - tail (int) – the id of the tail node
- head (int) – the id of the head node
Raises: gum.InvalidEdge– If arc.tail and/or arc.head are not in the ID.gum.InvalidEdge– If tail is a utility node
-
addChanceNode(self, variable, id=0)¶ addChanceNode(self, variable, aContent, id=0) -> int
Add a chance variable, it’s associate node and it’s CPT.
The id of the new variable is automatically generated.
Parameters: - variable (pyAgrum.DiscreteVariable) – the variable added by copy.
- id (int) – the chosen id. If 0, the NodeGraphPart will choose.
Warning
give an id (not 0) should be reserved for rare and specific situations !!!
Returns: the id of the added variable. Return type: int Raises: gum.DuplicateElement– If id(<>0) is already used
-
addDecisionNode(self, variable, id=0)¶ Add a decision variable.
The id of the new variable is automatically generated.
Parameters: - variable (pyAgrum.DiscreteVariable) – the variable added by copy.
- id (int) – the chosen id. If 0, the NodeGraphPart will choose.
Warning
give an id (not 0) should be reserved for rare and specific situations !!!
Returns: the id of the added variable. Return type: int Raises: gum.DuplicateElement– If id(<>0) is already used
-
addUtilityNode(self, variable, id=0)¶ addUtilityNode(self, variable, aContent, id=0) -> int
Add a utility variable, it’s associate node and it’s UT.
The id of the new variable is automatically generated.
Parameters: - variable (pyAgrum.DiscreteVariable) – the variable added by copy
- id (int) – the chosen id. If 0, the NodeGraphPart will choose
Warning
give an id (not 0) should be reserved for rare and specific situations !!!
Returns: the id of the added variable.
Return type: int
Raises: gum.InvalidArgument– If variable has more than one labelgum.DuplicateElement– If id(<>0) is already used
-
ancestors(self, norid)¶
-
arcs(self)¶ Returns: the list of all the arcs in the Influence Diagram. Return type: list
-
chanceNodeSize(self)¶ Returns: the number of chance nodes. Return type: int
-
changeVariableName(self, id, new_name)¶ changeVariableName(self, name, new_name)
Parameters: - id (int) – the node Id
- new_name (str) – the name of the variable
Raises: gum.DuplicateLabel– If this name already existsgum.NotFound– If no nodes matches id.
-
children(self, norid)¶ Parameters: id (int) – the id of the parent Returns: the set of all the children Return type: Set
-
clear(self)¶
-
completeInstantiation(self)¶
-
connectedComponents()¶ connected components from a graph/BN
Compute the connected components of a pyAgrum’s graph or Bayesian Network (more generally an object that has nodes, children/parents or neighbours methods)
The firstly visited node for each component is called a ‘root’ and is used as a key for the component. This root has been arbitrarily chosen during the algorithm.
Returns: dict of connected components (as set of nodeIds (int)) with a nodeId (root) of each component as key. Return type: dict(int,Set[int])
-
cpt(self, varId)¶ cpt(self, name) -> Potential
Returns the CPT of a variable.
Parameters: VarId (int) – A variable’s id in the pyAgrum.BayesNet. Returns: The variable’s CPT. Return type: pyAgrum.Potential Raises: gum.NotFound– If no variable’s id matches varId.
-
dag(self)¶ Returns: a constant reference to the dag of this BayesNet. Return type: pyAgrum.DAG
-
decisionNodeSize(self)¶ Returns: the number of decision nodes Return type: int
-
decisionOrder(self)¶
-
decisionOrderExists(self)¶ Returns: True if a directed path exist with all decision node Return type: bool
-
descendants(self, norid)¶
-
empty(self)¶
-
erase(self, id)¶ erase(self, name) erase(self, var)
Erase a Variable from the network and remove the variable from all his childs.
If no variable matches the id, then nothing is done.
Parameters: - id (int) – The id of the variable to erase.
- var (pyAgrum.DiscreteVariable) – The reference on the variable to remove.
-
eraseArc(self, arc)¶ eraseArc(self, tail, head) eraseArc(self, tail, head)
Removes an arc in the ID, and update diagram’s potential nodes cpt if necessary.
If (tail, head) doesn’t exist, the nothing happens.
Parameters: - arc (pyAgrum.Arc) – The arc to be removed.
- tail (int) – the id of the tail node
- head (int) – the id of the head node
-
exists(self, node)¶
-
existsArc(self, tail, head)¶ existsArc(self, nametail, namehead) -> bool
-
existsPathBetween(self, src, dest)¶ existsPathBetween(self, src, dest) -> bool
Returns: true if a path exists between two nodes. Return type: bool
-
family(self, norid)¶
-
static
fastPrototype(dotlike, domainSize=2)¶ - Create an Influence Diagram with a dot-like syntax which specifies:
- the structure ‘a->b<-c;b->d;c<-e;’.
- a prefix for the type of node (chance/decision/utiliy nodes):
- a : a chance node named ‘a’ (by default)
- $a : a utility node named ‘a’
- *a : a decision node named ‘a’
- the type of the variables with different syntax as postfix:
- by default, a variable is a gum.RangeVariable using the default domain size (second argument)
- with ‘a[10]’, the variable is a gum.RangeVariable using 10 as domain size (from 0 to 9)
- with ‘a[3,7]’, the variable is a gum.RangeVariable using a domainSize from 3 to 7
- with ‘a[1,3.14,5,6.2]’, the variable is a gum.DiscretizedVariable using the given ticks (at least 3 values)
- with ‘a{top|middle|bottom}’, the variable is a gum.LabelizedVariable using the given labels.
- with ‘a{-1|5|0|3}’, the variable is a gum.IntegerVariable using the sorted given values.
Note
- If the dot-like string contains such a specification more than once for a variable, the first specification will be used.
- the potentials (probabilities, utilities) are randomly generated.
- see also pyAgrum.fastID.
Examples
>>> import pyAgrum as gum >>> bn=gum.fastID('A->B[1,3]<-*C{yes|No}->$D<-E[1,2.5,3.9]',6)
Parameters: - dotlike (str) – the string containing the specification
- domainSize (int) – the default domain size for variables
Returns: the resulting Influence Diagram
Return type:
-
getDecisionGraph(self)¶ Returns: the temporal Graph. Return type: pyAgrum.DAG
-
hasSameStructure(self, other)¶ Parameters: pyAgrum.DAGmodel – a direct acyclic model Returns: True if all the named node are the same and all the named arcs are the same Return type: bool
-
idFromName(self, name)¶ Returns a variable’s id given its name.
Parameters: name (str) – the variable’s name from which the id is returned. Returns: the variable’s node id. Return type: int Raises: gum.NotFound– If no such name exists in the graph.
-
ids(self, names)¶
-
isChanceNode(self, varId)¶ isChanceNode(self, name) -> bool
Parameters: varId (int) – the tested node id. Returns: true if node is a chance node Return type: bool
-
isDecisionNode(self, varId)¶ isDecisionNode(self, name) -> bool
Parameters: varId (int) – the tested node id. Returns: true if node is a decision node Return type: bool
-
isIndependent(self, X, Y, Z)¶ isIndependent(self, X, Y, Z) -> bool isIndependent(self, Xname, Yname, Znames) -> bool isIndependent(self, Xnames, Ynames, Znames) -> bool
-
isUtilityNode(self, varId)¶ isUtilityNode(self, name) -> bool
Parameters: varId (int) – the tested node id. Returns: true if node is an utility node Return type: bool
-
loadBIFXML(self, name, l=(PyObject *) 0)¶ Load a BIFXML file.
Parameters: name (str) – the name’s file
Raises: gum.IOError– If file not foundgum.FatalError– If file is not valid
-
log10DomainSize(self)¶
-
moralGraph(self, clear=True)¶ Returns the moral graph of the BayesNet, formed by adding edges between all pairs of nodes that have a common child, and then making all edges in the graph undirected.
Returns: The moral graph Return type: pyAgrum.UndiGraph
-
moralizedAncestralGraph(self, nodes)¶
-
names(self)¶ Returns: The names of the InfluenceDiagram variables Return type: list
-
nodeId(self, var)¶ Parameters: var (pyAgrum.DiscreteVariable) – a variable Returns: the id of the variable Return type: int Raises: gum.IndexError– If the InfluenceDiagram does not contain the variable
-
nodes(self)¶ Returns: the set of ids Return type: set
-
nodeset(self, names)¶
-
parents(self, norid)¶ Parameters: id – The id of the child node Returns: the set of the parents ids. Return type: set
-
property(self, name)¶
-
propertyWithDefault(self, name, byDefault)¶
-
saveBIFXML(self, name)¶ Save the BayesNet in a BIFXML file.
Parameters: name (str) – the file’s name
-
setProperty(self, name, value)¶
-
size(self)¶ Returns: the number of nodes in the graph Return type: int
-
sizeArcs(self)¶ Returns: the number of arcs in the graph Return type: int
-
toDot(self)¶ Returns: a friendly display of the graph in DOT format Return type: str
-
topologicalOrder(self, clear=True)¶ Returns: the list of the nodes Ids in a topological order Return type: List Raises: gum.InvalidDirectedCycle– If this graph contains cycles
-
utility(self, varId)¶ utility(self, name) -> Potential
Parameters: varId (int) – the tested node id. Returns: the utility table of the node Return type: pyAgrum.Potential Raises: gum.IndexError– If the InfluenceDiagram does not contain the variable
-
utilityNodeSize(self)¶ Returns: the number of utility nodes Return type: int
-
variable(self, id)¶ variable(self, name) -> DiscreteVariable
Parameters: id (int) – the node id Returns: a constant reference over a variabe given it’s node id Return type: pyAgrum.DiscreteVariable Raises: gum.NotFound– If no variable’s id matches the parameter
-
variableFromName(self, name)¶ Parameters: name (str) – a variable’s name Returns: the variable Return type: pyAgrum.DiscreteVariable Raises: gum.IndexError– If the InfluenceDiagram does not contain the variable
-
variableNodeMap(self)¶
Inference¶
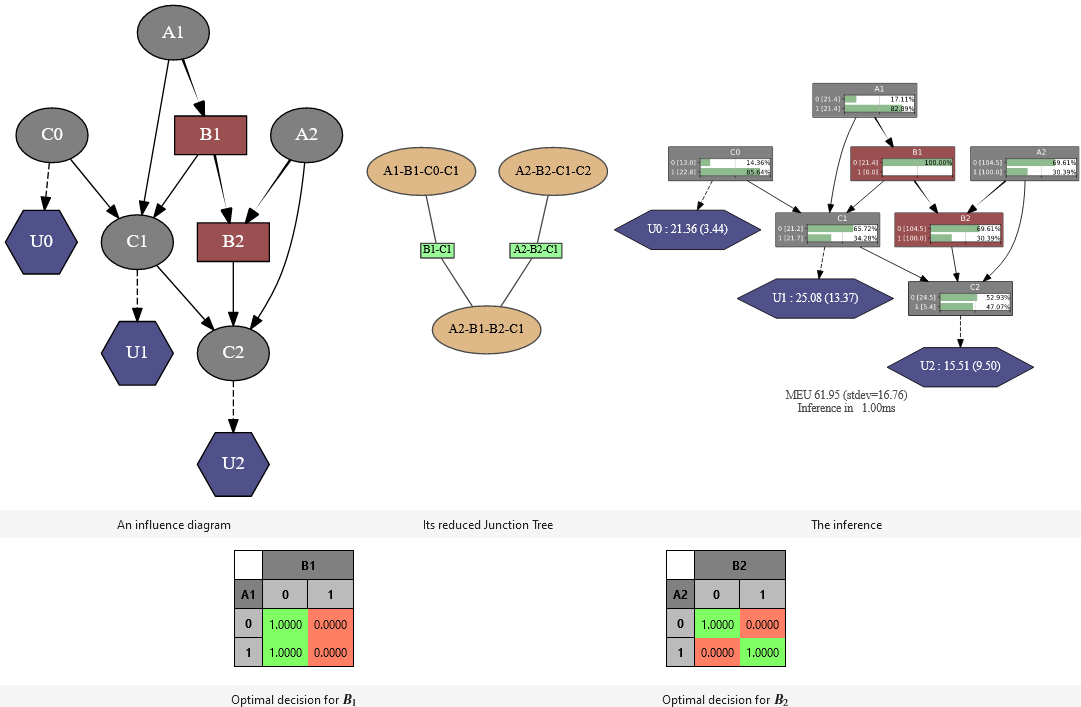
-
class
pyAgrum.ShaferShenoyLIMIDInference(infDiag: pyAgrum.InfluenceDiagram)¶ This inference considers the provided model as a LIMID rather than an influence diagram. It is an optimized implementation of the LIMID resolution algorithm. However an inference on a classical influence diagram can be performed by adding a assumption of the existence of the sequence of decision nodes to be solved, which also implies that the decision choices can have an impact on the rest of the sequence (Non Forgetting Assumption, cf. pyAgrum.ShaferShenoyLIMIDInference.addNoForgettingAssumption).
-
MEU(self)¶ MEU(self) -> PyObject *
Returns maximum expected utility obtained from inference.
Raises: gum.OperationNotAllowed– If no inference have yet been made
-
addEvidence(self, id, val)¶ addEvidence(self, nodeName, val) addEvidence(self, id, val) addEvidence(self, nodeName, val) addEvidence(self, id, vals) addEvidence(self, nodeName, vals)
-
addNoForgettingAssumption(self, ids)¶ addNoForgettingAssumption(self, names)
-
chgEvidence(self, id, val)¶ chgEvidence(self, nodeName, val) chgEvidence(self, id, val) chgEvidence(self, nodeName, val) chgEvidence(self, id, vals) chgEvidence(self, nodeName, vals)
-
clear(self)¶
-
eraseAllEvidence(self)¶ Removes all the evidence entered into the diagram.
-
eraseEvidence(self, id)¶ eraseEvidence(self, nodeName)
Parameters: evidence (pyAgrum.Potential) – the evidence to remove Raises: gum.IndexError– If the evidence does not belong to the influence diagram
-
hardEvidenceNodes(self)¶
-
hasEvidence(self, id)¶ hasEvidence(self, nodeName) -> bool
-
hasHardEvidence(self, nodeName)¶
-
hasNoForgettingAssumption(self)¶
-
hasSoftEvidence(self, id)¶ hasSoftEvidence(self, nodeName) -> bool
-
influenceDiagram(self)¶ Returns a constant reference over the InfluenceDiagram on which this class work.
Returns: the InfluenceDiagram on which this class work Return type: pyAgrum.InfluenceDiagram
-
isSolvable(self)¶
-
junctionTree(self)¶
-
makeInference(self)¶ Makes the inference.
-
meanVar(self, node)¶ meanVar(self, name) -> pair< double,double > meanVar(self, node) -> PyObject *
-
nbrEvidence(self)¶
-
nbrHardEvidence(self)¶
-
nbrSoftEvidence(self)¶
-
optimalDecision(self, decisionId)¶ optimalDecision(self, decisionName) -> Potential
Returns best choice for decision variable given in parameter ( based upon MEU criteria )
Parameters: decisionId (int,str) – the id or name of the decision variable
Raises: gum.OperationNotAllowed– If no inference have yet been madegum.InvalidNode– If node given in parmaeter is not a decision node
-
posterior(self, node)¶ posterior(self, name) -> Potential posterior(self, var) -> Potential posterior(self, nodeName) -> Potential
-
posteriorUtility(self, node)¶ posteriorUtility(self, name) -> Potential
-
reducedGraph(self)¶
-
reducedLIMID(self)¶
-
reversePartialOrder(self)¶
-
setEvidence(evidces)¶ Erase all the evidences and apply addEvidence(key,value) for every pairs in evidces.
Parameters: evidces (dict) – a dict of evidences
Raises: gum.InvalidArgument– If one value is not a value for the nodegum.InvalidArgument– If the size of a value is different from the domain side of the nodegum.FatalError– If one value is a vector of 0sgum.UndefinedElement– If one node does not belong to the influence diagram
-
softEvidenceNodes(self)¶
-
updateEvidence(evidces)¶ Apply chgEvidence(key,value) for every pairs in evidces (or addEvidence).
Parameters: evidces (dict) – a dict of evidences
Raises: gum.InvalidArgument– If one value is not a value for the nodegum.InvalidArgument– If the size of a value is different from the domain side of the nodegum.FatalError– If one value is a vector of 0sgum.UndefinedElement– If one node does not belong to the Bayesian network
-