Coloring and exporting graphical models as image (pdf, png)
In [1]:
from pylab import *
import matplotlib.pyplot as plt
In [2]:
import pyAgrum as gum
import pyAgrum.lib.notebook as gnb
In [3]:
bn=gum.fastBN("a->b->c->d;b->e->d->f;g->c")
gnb.flow.row(bn,gnb.getInference(bn))
customizing colours and width for model and inference
In [4]:
def nodevalue(n):
return 0.5 if n in "aeiou" else 0.7
def arcvalue(a):
return (10-a[0])*a[1]
def arcvalue2(a):
return (a[0]+a[1]+5)/22
gnb.showBN(bn,
nodeColor={n:nodevalue(n) for n in bn.names()},
arcWidth={a:arcvalue(a) for a in bn.arcs()},
arcLabel={a:f"v={arcvalue(a):02d}" for a in bn.arcs()},
arcColor={a:arcvalue2(a) for a in bn.arcs()})
In [5]:
gnb.showInference(bn,
targets={"a","g","f","b"},
evs={'e':0},
nodeColor={n:nodevalue(n) for n in bn.names()},
arcWidth={a:arcvalue(a) for a in bn.arcs()})
In [6]:
gnb.flow.row(gnb.getBN(bn,
nodeColor={n:nodevalue(n) for n in bn.names()},
arcWidth={a:arcvalue(a) for a in bn.arcs()}),
gnb.getInference(bn,
nodeColor={n:nodevalue(n) for n in bn.names()},
arcWidth={a:arcvalue(a) for a in bn.arcs()})
)
In [7]:
import matplotlib.pyplot as plt
mycmap=plt.get_cmap('Reds')
formyarcs=plt.get_cmap('winter')
gnb.flow.row(gnb.getBN(bn,
nodeColor={n:nodevalue(n) for n in bn.names()},
arcColor={a:arcvalue2(a) for a in bn.arcs()},
cmapNode=mycmap,
cmapArc=formyarcs),
gnb.getInference(bn,
nodeColor={n:nodevalue(n) for n in bn.names()},
arcColor={a:arcvalue2(a) for a in bn.arcs()},
arcWidth={a:arcvalue(a) for a in bn.arcs()},
cmapNode=mycmap,
cmapArc=formyarcs)
)
Exporting model and inference as image
Exporting as image (pdf, png, etc.) has been gathered in 2 functions : pyAgrum.lib.image.export() and pyAgrum.lib.image.exportInference(). The argument are the same as for pyAgrum.notebook.show{Model} and pyAgrum.notebook.show{Inference}.
In [8]:
import pyAgrum.lib.image as gumimage
from IPython.display import Image # to display the exported images
In [9]:
gumimage.export(bn,"out/test_export.png")
Image(filename='out/test_export.png')
Out[9]:
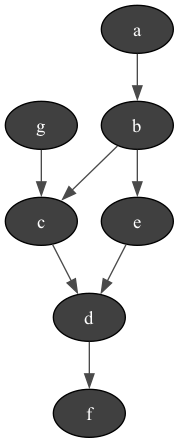
In [10]:
bn = gum.fastBN("a->b->d;a->c->d[3]->e;f->b")
gumimage.export(bn,"out/test_export.png",
nodeColor={'a': 1,
'b': 0.3,
'c': 0.4,
'd': 0.1,
'e': 0.2,
'f': 0.5},
arcColor={(0, 1): 0.2,
(1, 2): 0.5},
arcWidth={(0, 3): 0.4,
(3, 2): 0.5,
(2,4) :0.6})
Image(filename='out/test_export.png')
Out[10]:
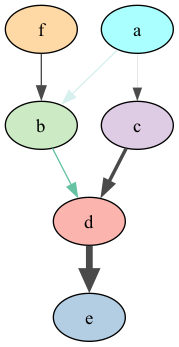
In [11]:
gumimage.exportInference(bn,"out/test_export.png")
Image(filename='out/test_export.png')
Out[11]:
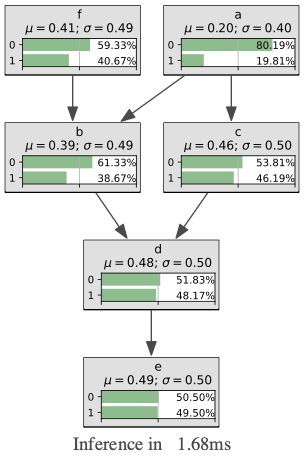
In [12]:
gumimage.export(bn,"out/test_export.pdf")
Link to out/test_export.pdf
exporting inference with evidence
In [13]:
bn=gum.loadBN("res/alarm.dsl")
gumimage.exportInference(bn,"out/test_export.pdf",
evs={"CO":1,"VENTLUNG":1},
targets={"VENTALV",
"CATECHOL",
"HR",
"MINVOLSET",
"ANAPHYLAXIS",
"STROKEVOLUME",
"ERRLOWOUTPUT",
"HBR",
"PULMEMBOLUS",
"HISTORY",
"BP",
"PRESS",
"CO"},
size="15!")
Link to out/test_export.pdf
Other models
Other models can also use these functions.
In [14]:
infdiag=gum.loadID("res/OilWildcatter.bifxml")
gumimage.export(infdiag,"out/test_export.pdf")
Link to out/test_export.pdf
In [15]:
gumimage.exportInference(infdiag,"out/test_export.pdf")
Link to out/test_export.pdf
Exporting any object with toDot() method
In [16]:
import pyAgrum.causal as csl
obs1 = gum.fastBN("Smoking->Cancer")
modele3 = csl.CausalModel(obs1, [("Genotype", ["Smoking","Cancer"])], True)
gumimage.export(modele3,"out/test_export.png") # a causal model has a toDot method.
Image(filename='out/test_export.png')
Out[16]:
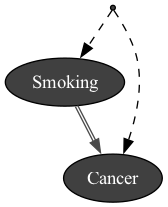
In [17]:
bn=gum.fastBN("a->b->c->d;b->e->d->f;g->c")
ie=gum.LazyPropagation(bn)
jt=ie.junctionTree()
gumimage.export(jt,"out/test_export.png") # a JunctionTree has a method jt.toDot()
Image(filename='out/test_export.png')
Out[17]:
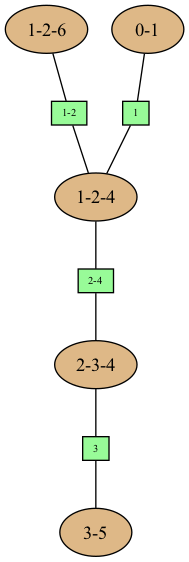
… or even a string in dot syntax
In [18]:
gumimage.export(jt.toDotWithNames(bn),"out/test_export.png") # jt.toDotWithNames(bn) creates a dot-string for a junction tree with names of variables
Image(filename='out/test_export.png')
Out[18]:
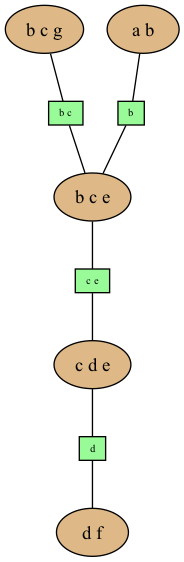
Exporting to pyplot
In [19]:
import matplotlib.pyplot as plt
bn=gum.fastBN("A->B->C<-D")
plt.imshow(gumimage.export(bn))
plt.show()
plt.imshow(gumimage.exportInference(bn,size="15!"))
plt.show()
plt.figure(figsize = (10,10))
plt.imshow(gumimage.exportInference(bn,size="15!"))
plt.show()

