Model for Bayesian Network
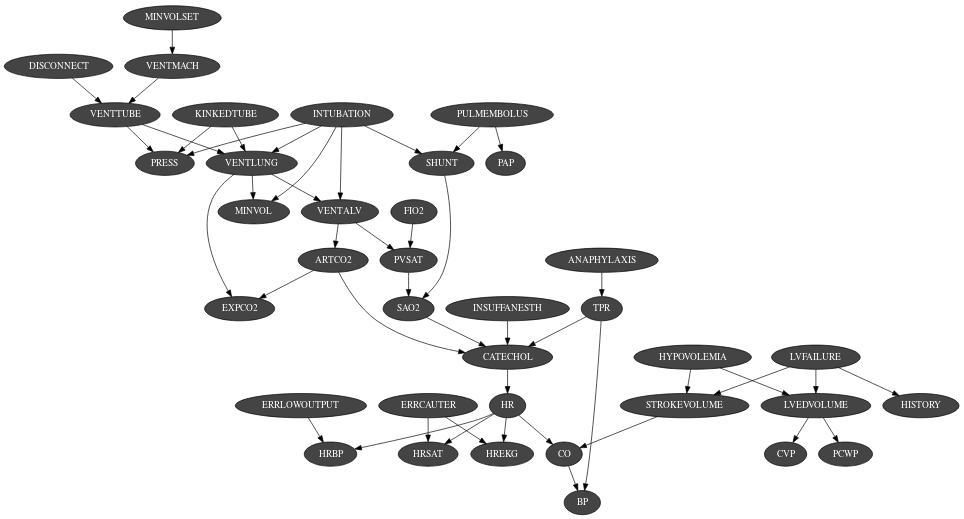
The Bayesian network is the main graphical model of pyAgrum. A Bayesian network is a directed probabilistic graphical model based on a DAG. It represents a joint distribution over a set of random variables. In pyAgrum, the variables are (for now) only discrete.
A Bayesian network uses a directed acyclic graph (DAG) to represent conditional independence in the joint distribution. These conditional independence allow to factorize the joint distribution, thereby allowing to compactly represent very large ones.
Moreover, inference algorithms can also use this graph to speed up the computations. Finally, the Bayesian networks can be learnt from data.
Tutorial
- class pyAgrum.BayesNet(*args)
BayesNet represents a Bayesian network.
- BayesNet(name=’’) -> BayesNet
- Parameters:
name (str) – the name of the Bayes Net
- BayesNet(source) -> BayesNet
- Parameters:
source (pyAgrum.BayesNet) – the Bayesian network to copy
- add(*args)
Add a variable to the pyAgrum.BayesNet.
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable added
descr (str) – the description of the variable (following fast syntax)
nbrmod (int) – the number of modalities for the new variable
id (int) – the variable forced id in the pyAgrum.BayesNet
- Returns:
the id of the new node
- Return type:
int
- Raises:
pyAgrum.DuplicateLabel – If variable.name() or id is already used in this pyAgrum.BayesNet.
pyAgrum.NotAllowed – If nbrmod is less than 2
- addAMPLITUDE(var)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –
- Returns:
the id of the added value
- Return type:
int
- addAND(var)
Add a variable, it’s associate node and an AND implementation.
The id of the new variable is automatically generated.
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy.
var (
DiscreteVariable) –
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.SizeError – If variable.domainSize()>2
- addArc(*args)
Add an arc in the BN, and update arc.head’s CPT.
- Parameters:
head (Union[int,str]) – a variable’s id (int) or name
head – a variable’s id (int) or name
- Raises:
pyAgrum.InvalidEdge – If arc.tail and/or arc.head are not in the BN.
pyAgrum.DuplicateElement – If the arc already exists.
- Return type:
None
- addArcs(listArcs)
add a list of arcs in te model.
- Parameters:
listArcs (List[Tuple[intstr,intstr]]) – the list of arcs
- addCOUNT(var, value=1)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –value (
int) –
- Returns:
the id of the added value
- Return type:
int
- addEXISTS(var, value=1)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –value (
int) –
- Returns:
the id of the added value
- Return type:
int
- addFORALL(var, value=1)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –value (
int) –
- Returns:
the id of the added variable.
- Return type:
int
- addLogit(*args)
Add a variable, its associate node and a Logit implementation.
(The id of the new variable can be automatically generated.)
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
externalWeight (float) – the added external weight
id (int) – The proposed id for the variable.
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.DuplicateElement – If id is already used
- addMAX(var)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –
- Returns:
the id of the added value
- Return type:
int
- addMEDIAN(var)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –
- Returns:
the id of the added value
- Return type:
int
- addMIN(var)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –
- Returns:
the id of the added value
- Return type:
int
- addNoisyAND(*args)
Add a variable, its associate node and a noisyAND implementation.
(The id of the new variable can be automatically generated.)
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
externalWeight (float) – the added external weight
id (int) – The proposed id for the variable.
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.DuplicateElement – If id is already used
- addNoisyOR(*args)
Add a variable, it’s associate node and a noisyOR implementation.
Since it seems that the ‘classical’ noisyOR is the Compound noisyOR, we keep the addNoisyOR as an alias for addNoisyORCompound.
(The id of the new variable can be automatically generated.)
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
externalWeight (float) – the added external weight
id (int) – The proposed id for the variable.
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.DuplicateElement – If id is already used
- addNoisyORCompound(*args)
Add a variable, it’s associate node and a noisyOR implementation.
Since it seems that the ‘classical’ noisyOR is the Compound noisyOR, we keep the addNoisyOR as an alias for addNoisyORCompound.
(The id of the new variable can be automatically generated.)
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
externalWeight (float) – the added external weight
id (int) – The proposed id for the variable.
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.DuplicateElement – If id is already used
- addNoisyORNet(*args)
Add a variable, its associate node and a noisyOR implementation.
Since it seems that the ‘classical’ noisyOR is the Compound noisyOR, we keep the addNoisyOR as an alias for addNoisyORCompound.
(The id of the new variable can be automatically generated.)
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
externalWeight (float) – the added external weight
id (int) – The proposed id for the variable.
- Returns:
the id of the added variable.
- Return type:
int
- addOR(var)
Add a variable, it’s associate node and an OR implementation.
The id of the new variable is automatically generated.
Warning
If parents are not boolean, all value>1 is True
- Parameters:
variable (pyAgrum.DiscreteVariable) – The variable added by copy
var (
DiscreteVariable) –
- Returns:
the id of the added variable.
- Return type:
int
- Raises:
pyAgrum.SizeError – If variable.domainSize()>2
- addSUM(var)
Others aggregators
- Parameters:
variable (pyAgrum.DiscreteVariable) – the variable to be added
var (
DiscreteVariable) –
- Returns:
the id of the added value
- Return type:
int
- addStructureListener(whenNodeAdded=None, whenNodeDeleted=None, whenArcAdded=None, whenArcDeleted=None)
Add the listeners in parameters to the list of existing ones.
- Parameters:
whenNodeAdded (lambda expression) – a function for when a node is added
whenNodeDeleted (lambda expression) – a function for when a node is removed
whenArcAdded (lambda expression) – a function for when an arc is added
whenArcDeleted (lambda expression) – a function for when an arc is removed
- addVariables(listFastVariables, default_nbr_mod=2)
Add a list of variable in the form of ‘fast’ syntax.
- Parameters:
listFastVariables (List[str]) – the list of variables in ‘fast’ syntax.
default_nbr_mod (int) – the number of modalities for the variable if not specified following fast syntax. Note that default_nbr_mod=1 is mandatory to create variables with only one modality (for utility for instance).
- Returns:
the list of created ids.
- Return type:
List[int]
- addWeightedArc(*args)
Add an arc in the BN, and update arc.head’s CPT.
- Parameters:
head (Union[int,str]) – a variable’s id (int) or name
tail (Union[int,str]) – a variable’s id (int) or name
causalWeight (float) – the added causal weight
- Raises:
pyAgrum.InvalidArc – If arc.tail and/or arc.head are not in the BN.
pyAgrum.InvalidArc – If variable in arc.head is not a NoisyOR variable.
- Return type:
None
- adjacencyMatrix()
adjacency matrix from a graph/graphical models
Compute the adjacency matrix of a pyAgrum’s graph or graphical models (more generally an object that has nodes, children/parents or neighbours methods)
- Returns:
adjacency matrix (as numpy.ndarray) with nodeId as key.
- Return type:
numpy.ndarray
- ancestors(norid)
give the set of nodeid of ancestors of a node
- Parameters:
norid (str|int) – the name or the id of the node
- Returns:
the set of ids of the ancestors of node norid.
- Return type:
Set[int]
- arcs()
- Returns:
The lisf of arcs in the IBayesNet
- Return type:
list
- beginTopologyTransformation()
When inserting/removing arcs, node CPTs change their dimension with a cost in time. begin Multiple Change for all CPTs These functions delay the CPTs change to be done just once at the end of a sequence of topology modification, begins a sequence of insertions/deletions of arcs without changing the dimensions of the CPTs.
- Return type:
None
- changePotential(*args)
change the CPT associated to nodeId to newPot delete the old CPT associated to nodeId.
- Parameters:
var (Union[int,str]) – the current name or the id of the variable
newPot (pyAgrum.Potential) – the new potential
- Raises:
pyAgrum.NotAllowed – If newPot has not the same signature as __probaMap[NodeId]
- Return type:
None
- changeVariableLabel(*args)
change the label of the variable associated to nodeId to the new value.
- Parameters:
var (Union[int,str]) – the current name or the id of the variable
old_label (str) – the new label
new_label (str) – the new label
- Raises:
pyAgrum.NotFound – if id/name is not a variable or if old_label does not exist.
- Return type:
None
- changeVariableName(*args)
Changes a variable’s name in the pyAgrum.BayesNet.
This will change the “pyAgrum.DiscreteVariable” names in the pyAgrum.BayesNet.
- Parameters:
var (Union[int,str]) – the current name or the id of the variable
new_name (str) – the new name of the variable
- Raises:
pyAgrum.DuplicateLabel – If new_name is already used in this BayesNet.
pyAgrum.NotFound – If no variable matches id.
- Return type:
None
- check()
Check if the BayesNet is consistent (variables, CPT, …)
- Returns:
list of found issues
- Return type:
List[str]
- children(norid)
- Parameters:
id (int) – the id of the parent
norid (
object) –
- Returns:
the set of all the children
- Return type:
Set
- clear()
Clear the whole BayesNet
- Return type:
None
- completeInstantiation()
Give an instantiation over all the variables of the model
- Returns:
a complete Instantiation for the model
- Return type:
- connectedComponents()
connected components from a graph/graphical models
Compute the connected components of a pyAgrum’s graph or graphical models (more generally an object that has nodes, children/parents or neighbours methods)
The firstly visited node for each component is called a ‘root’ and is used as a key for the component. This root has been arbitrarily chosen during the algorithm.
- Returns:
dict of connected components (as set of nodeIds (int)) with a nodeId (root) of each component as key.
- Return type:
dict(int,Set[int])
- cpt(*args)
Returns the CPT of a variable.
- Parameters:
VarId (Union[int,str]) – a variable’s id (int) or name
- Returns:
The variable’s CPT.
- Return type:
- Raises:
pyAgrum.NotFound – If no variable’s id matches varId.
- dag()
- Returns:
a constant reference to the dag of this BayesNet.
- Return type:
- descendants(norid)
give the set of nodeid of descendants of a node
- Parameters:
norid (str|int) – the name or the id of the node
- Returns:
the set of ids of the descendants of node norid.
- Return type:
Set[int]
- dim()
Returns the dimension (the number of free parameters) in this BayesNet.
- Returns:
the dimension of the BayesNet
- Return type:
int
- empty()
Check if there are some variables in the model.
- Returns:
True if there is no variable in the model.
- Return type:
bool
- endTopologyTransformation()
Terminates a sequence of insertions/deletions of arcs by adjusting all CPTs dimensions. End Multiple Change for all CPTs.
- Return type:
- erase(*args)
Remove a variable from the pyAgrum.BayesNet.
Removes the corresponding variable from the pyAgrum.BayesNet and from all of it’s children pyAgrum.Potential.
If no variable matches the given id, then nothing is done.
- Parameters:
var (Union[int,str,pyAgrum.DiscreteVariable]) – the current name, the id of the variable or a reference to the variable
- Return type:
None
- eraseArc(*args)
Removes an arc in the BN, and update head’s CTP.
If (tail, head) doesn’t exist, the nothing happens.
- Parameters:
arc (pyAgrum.Arc when calling eraseArc(arc)) – The arc to be removed.
head (Union[int,str]) – a variable’s id (int) or name for the head when calling eraseArc(head,tail)
tail (Union[int,str]) – a variable’s id (int) or name for the tail when calling eraseArc(head,tail)
- Return type:
None
- evIn(name, val1, val2)
- Parameters:
name (
str) –val1 (
float) –val2 (
float) –
- Return type:
- exists(*args)
Check if a node with this name or id exists
- Parameters:
norid (str|int) – name or id of the searched node
- Returns:
True if there is a node with such a name or id
- Return type:
bool
- existsArc(*args)
Check if an arc exists
- Parameters:
tail (str|int) – the name or id of the tail of the arc
head (str|int) – the name or the id of the head of the arc
- Returns:
True if tail->head is an arc.
- Return type:
bool
- family(norid)
give the set of parents of a node and the node
- Parameters:
norid (str|int) – the node
- Returns:
the set of nodeId of the family of the node norid
- Return type:
Set[int]
- static fastPrototype(*args)
- Create a Bayesian network with a dot-like syntax which specifies:
the structure ‘a->b->c;b->d<-e;’.
the type of the variables with different syntax:
by default, a variable is a pyAgrum.RangeVariable using the default domain size ([2])
with ‘a[10]’, the variable is a pyAgrum.RangeVariable using 10 as domain size (from 0 to 9)
with ‘a[3,7]’, the variable is a pyAgrum.RangeVariable using a domainSize from 3 to 7
with ‘a[1,3.14,5,6.2]’, the variable is a pyAgrum.DiscretizedVariable using the given ticks (at least 3 values)
with ‘a{top|middle|bottom}’, the variable is a pyAgrum.LabelizedVariable using the given labels.
with ‘a{-1|5|0|3}’, the variable is a pyAgrum.IntegerVariable using the sorted given values.
with ‘a{-0.5|5.01|0|3.1415}’, the variable is a pyAgrum.NumericalDiscreteVariable using the sorted given values.
Note
If the dot-like string contains such a specification more than once for a variable, the first specification will be used.
the CPTs are randomly generated.
see also pyAgrum.fastBN.
Examples
>>> import pyAgrum as gum >>> bn=pyAgrum.BayesNet.fastPrototype('A->B[1,3]<-C{yes|No}->D[2,4]<-E[1,2.5,3.9]',6)
- Parameters:
dotlike (str) – the string containing the specification
domainSize (int or str) – the default domain size or the default domain for variables
- Returns:
the resulting Bayesian network
- Return type:
- generateCPT(*args)
Randomly generate CPT for a given node in a given structure.
- Parameters:
node (Union[int,str]) – a variable’s id (int) or name
- Return type:
None
- generateCPTs()
Randomly generates CPTs for a given structure.
- Return type:
None
- hasSameStructure(other)
- Parameters:
pyAgrum.DAGmodel – a direct acyclic model
- Returns:
True if all the named node are the same and all the named arcs are the same
- Return type:
bool
- idFromName(name)
Returns a variable’s id given its name in the graph.
- Parameters:
name (str) – The variable’s name from which the id is returned.
Notes
A convenient shortcut for g.variableFromName(name) is g[name].
- Returns:
The variable’s node id.
- Return type:
int
- Raises:
pyAgrum.NotFound – If name does not match a variable in the graph
- ids(names)
List of ids for a list of names of variables in the model
- Parameters:
lov (List[str]) – List of variable names
names (
List[str]) –
- Returns:
The ids for the list of names of the graph variables
- Return type:
List[int]
- isIndependent(*args)
check if nodes X and nodes Y are independent given nodes Z
- Parameters:
X (str|intList[str|int]) – a list of of nodeIds or names
Y (str|intList[str|int]) – a list of of nodeIds or names
Z (str|intList[str|int]) – a list of of nodeIds or names
- Raises:
InvalidArgument – if X and Y share variables
- Returns:
True if X and Y are independent given Z in the model
- Return type:
bool
- jointProbability(i)
- Parameters:
i (pyAgrum.instantiation) – an instantiation of the variables
- Returns:
a parameter of the joint probability for the BayesNet
- Return type:
float
Warning
a variable not present in the instantiation is assumed to be instantiated to 0
- loadBIF(name, l=None)
Load a BIF file.
- Parameters:
name (str) – the file’s name
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadBIFXML(name, l=None)
Load a BIFXML file.
- Parameters:
name (str) – the name’s file
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadDSL(name, l=None)
Load a DSL file.
- Parameters:
name (str) – the file’s name
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadNET(name, l=None)
Load a NET file.
- Parameters:
name (str) – the name’s file
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadO3PRM(*args)
Load an O3PRM file.
Warning
The O3PRM language is the only language allowing to manipulate not only DiscretizedVariable but also RangeVariable and LabelizedVariable.
- Parameters:
name (str) – the file’s name
system (str) – the system’s name
classpath (str) – the classpath
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadUAI(name, l=None)
Load an UAI file.
- Parameters:
name (str) – the name’s file
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- loadXDSL(name, l=None)
Load a XDSL file.
- Parameters:
name (str) – the file’s name
l (list) – list of functions to execute
- Raises:
pyAgrum.IOError – If file not found
pyAgrum.FatalError – If file is not valid
- Return type:
str
- log10DomainSize()
returns the log10 of the domain size of the model defined as the product of the domain sizes of the variables in the model.
- Returns:
the log10 domain size.
- Return type:
float
- log2JointProbability(i)
- Parameters:
i (pyAgrum.instantiation) – an instantiation of the variables
- Returns:
a parameter of the log joint probability for the BayesNet
- Return type:
float
Warning
a variable not present in the instantiation is assumed to be instantiated to 0
- maxNonOneParam()
- Returns:
The biggest value (not equal to 1) in the CPTs of the BayesNet
- Return type:
float
- maxParam()
- Returns:
the biggest value in the CPTs of the BayesNet
- Return type:
float
- maxVarDomainSize()
- Returns:
the biggest domain size among the variables of the BayesNet
- Return type:
int
- minNonZeroParam()
- Returns:
the smallest value (not equal to 0) in the CPTs of the IBayesNet
- Return type:
float
- minParam()
- Returns:
the smallest value in the CPTs of the IBayesNet
- Return type:
float
- minimalCondSet(*args)
Returns, given one or many targets and a list of variables, the minimal set of those needed to calculate the target/targets.
- Parameters:
target (int) – The id of the target
targets (List[int]) – The ids of the targets
list (List[int]) – The list of available variables
- Returns:
The minimal set of variables
- Return type:
Set[int]
- moralGraph()
Returns the moral graph of the BayesNet, formed by adding edges between all pairs of nodes that have a common child, and then making all edges in the graph undirected.
- Returns:
The moral graph
- Return type:
- moralizedAncestralGraph(nodes)
build a UndiGraph by moralizing the Ancestral Graph of a list of nodes
- Parameters:
nodes (str|intList[str|int]) – the list of of nodeIds or names
Warning
pyAgrum.UndiGraph only knows NodeId. Hence the moralized ancestral graph does not include the names of the variables.graph
- Returns:
the moralized ancestral graph of the nodes
- Return type:
- names()
Set of names of variables in the model
- Returns:
The names of the graph variables
- Return type:
Set[str]
- nodeId(var)
- Parameters:
var (pyAgrum.DiscreteVariable) – a variable
- Returns:
the id of the variable
- Return type:
int
- Raises:
pyAgrum.IndexError – If the graph does not contain the variable
- nodes()
- Returns:
the set of ids
- Return type:
Set[int]
- nodeset(names)
Set of ids for a list of names of variables in the model
- Parameters:
lov (List[str]) – List of variable names
names (
List[str]) –
- Returns:
The set of ids for the list of names of the graph variables
- Return type:
Set[int]
- parents(norid)
- Parameters:
id – The id of the child node
norid (
object) –
- Returns:
the set of the parents ids.
- Return type:
Set
- properties()
- Return type:
List[str]
- reverseArc(*args)
Reverses an arc while preserving the same joint distribution.
- Parameters:
tail – (int) the id of the tail variable
head – (int) the id of the head variable
tail – (str) the name of the tail variable
head – (str) the name of the head variable
arc (pyAgrum.Arc) – an arc
- Raises:
pyAgrum.InvalidArc – If the arc does not exsit or if its reversal would induce a directed cycle.
- Return type:
None
- saveBIF(name, allowModificationWhenSaving=False)
Save the BayesNet in a BIF file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveBIFXML(name, allowModificationWhenSaving=False)
Save the BayesNet in a BIFXML file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveDSL(name, allowModificationWhenSaving=False)
Save the BayesNet in a DSL file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveNET(name, allowModificationWhenSaving=False)
Save the BayesNet in a NET file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveO3PRM(name, allowModificationWhenSaving=False)
Save the BayesNet in an O3PRM file.
Warning
The O3PRM language is the only language allowing to manipulate not only DiscretizedVariable but also RangeVariable and LabelizedVariable.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveUAI(name, allowModificationWhenSaving=False)
Save the BayesNet in an UAI file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – False by default. if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- saveXDSL(name, allowModificationWhenSaving=False)
Save the BayesNet in a XDSL file.
- Parameters:
name (str) – the file’s name
allowModificationWhenSaving (bool) – (not used). if true, syntax errors are corrected when saving the file. If false, they throw a FatalError.
- Return type:
None
- size()
- Returns:
the number of nodes in the graph
- Return type:
int
- sizeArcs()
- Returns:
the number of arcs in the graph
- Return type:
int
- property thisown
The membership flag
- toDot()
- Returns:
a friendly display of the graph in DOT format
- Return type:
str
- topologicalOrder()
- Returns:
the list of the nodes Ids in a topological order
- Return type:
List
- Raises:
pyAgrum.InvalidDirectedCycle – If this graph contains cycles
- variable(*args)
- Parameters:
id (int) – a variable’s id
name (str) – a variable’s name
- Returns:
the variable
- Return type:
- Raises:
pyAgrum.IndexError – If the graph does not contain the variable
- variableFromName(name)
- Parameters:
name (str) – a variable’s name
- Returns:
the variable
- Return type:
- Raises:
pyAgrum.IndexError – If the graph does not contain the variable
- variableNodeMap()
- Returns:
the variable node map
- Return type:
pyAgrum.variableNodeMap