Module dynamic bayesian network¶
Basic implementation for dynamic Bayesian Networks in pyAgrum
-
pyAgrum.lib.dynamicBN.getTimeSlices(dbn, size=None) Try to correctly represent dBN and 2TBN as an HTML string
Parameters: - dbn – the dynamic BN
- size – size of the figue
- format – png/svg
-
pyAgrum.lib.dynamicBN.getTimeSlicesRange(dbn) get the range and (name,radical) of each variables
Parameters: dbn – a 2TBN or an unrolled BN Returns: all the timeslice of a dbn e.g. [‘0’,’t’] for a classic 2TBN range(T) for a classic unrolled BN
-
pyAgrum.lib.dynamicBN.is2TBN(bn)
-
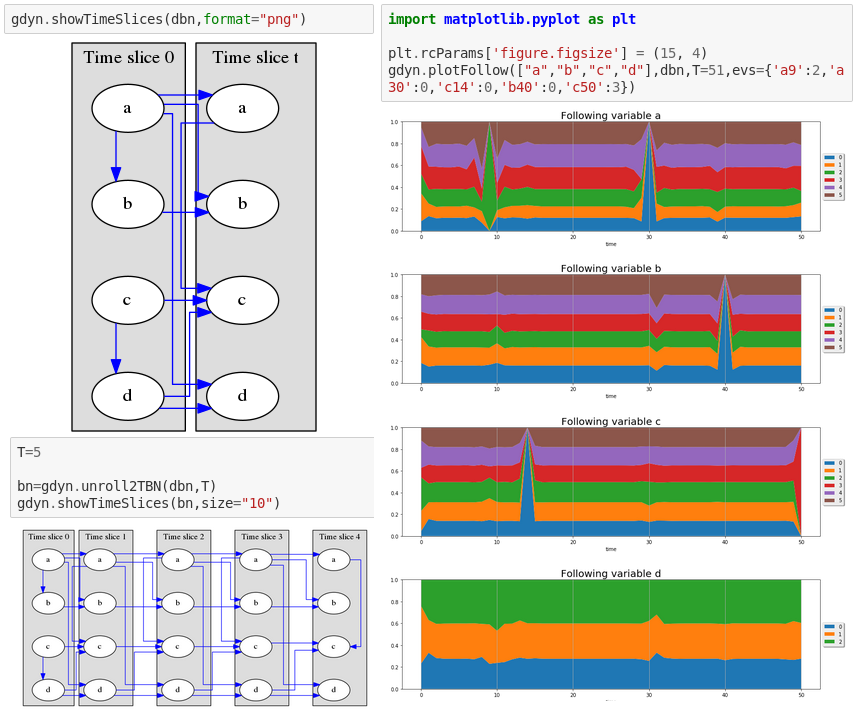
pyAgrum.lib.dynamicBN.plotFollow(lovars, twoTdbn, T, evs) plots modifications of variables in a 2TDN knowing the size of the time window (T) and the evidence on the sequence.
Parameters: - lovars – list of variables to follow
- twoTdbn – the two-timeslice dbn
- T – the time range
- evs – observations
-
pyAgrum.lib.dynamicBN.plotFollowUnrolled(lovars, dbn, T, evs) plot the dynamic evolution of a list of vars with a dBN
Parameters: - lovars – list of variables to follow
- dbn – the unrolled dbn
- T – the time range
- evs – observations
-
pyAgrum.lib.dynamicBN.realNameFrom2TBNname(name, ts) @return dynamic name from static name and timeslice (no check)
-
pyAgrum.lib.dynamicBN.showTimeSlices(dbn, size=None) Try to correctly display dBN and 2TBN
Parameters: - dbn – the dynamic BN
- size – size of the figue
- format – png/svg
-
pyAgrum.lib.dynamicBN.unroll2TBN(dbn, nbr) unroll a 2TBN given the nbr of timeslices
Parameters: - dbn – the dBN
- nbr – the number of timeslice
Returns: unrolled BN from a 2TBN and the nbr of timeslices